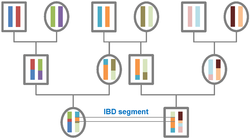
Identity by descent, known commonly as IBD, is a term employed in the field of genetic genealogy to refer to a segment of DNA that is shared by two or more individuals and has been passed down from a shared ancestor without any intervening recombination.[1][2] This concept is fundamental to understanding genetically mediated similarities among relatives and plays a crucial role in genealogical research and family history studies via DNA testing. Gene identity by descent (IBD) underlies genetically mediated similarities among relatives. It is traced through ancestral meiosis and is defined relative to founders of a pedigree, or to some time point or mutational origin in the coalescent of a set of extant genes in a population.[3] The random process underlying changes in the patterns of IBD across the genome is recombination, so the natural context for defining IBD is the ancestral recombination graph (ARG), which specifies the complete ancestry of a collection of chromosomes.
Research your ancestors on MyHeritage
Differences between IBD and IBS
Identity by descent (IBD) refers to alleles that are descended from a common ancestor in a base population.[4] Identity by state (IBS) simply refers to alleles that are the same, irrespective of whether they are inherited from a recent ancestor. While IBD segments are always IBS, segments that are not IBD can still be IBS due to the same mutations in different individuals or recombinations that do not alter the segment.
Identification of IDB segments
The identification of IBD segments can be done by comparing the DNA of two or more individuals to find matching segments that have been inherited from a common ancestor.[1] These segments are considered to match if all the alleles on a paternal or maternal chromosome are identical, with the exception of rare mutations and genotyping errors, and if the minimum threshold conditions set by the testing provider have been met. The length of IBD segments can be measured in centiMorgans (cM), a unit of genetic distance, or in megabases (Mb), a unit of physical distance.[2] The length and frequency of an IBD segment can provide valuable information about the relationship between individuals sharing the segment. Generally, larger segments indicate a closer relationship, but the frequency of the segment must also be considered. High-frequency IBD segments are more likely to signal distant sharing at the population level, while a segment observed in only two independently sampled individuals is more likely to be IBD.[5]
Methods and software for detection of IBD segments
Several programs have been developed for detecting IBD segments in unrelated individuals, including GERMLINE, fastIBD, and ISCA. These programs use different algorithms and approaches to identify IBD segments, with varying levels of accuracy and computational efficiency. Some programs, such as GERMLINE, focus on detecting long IBD segments, while others, like fastIBD, prioritize the detection of shorter segments.[6]
Applications of IDB in genealogy
DNA segments that are inherited by descent play a significant role in genealogical research and family history studies. By identifying shared IBD segments among individuals with a DNA test like MyHeritage DNA, researchers can determine relationships between relatives and trace their common ancestors,[1] as well as gain insights into the demographic history of various populations, including bottlenecks and admixture events.[5] This information can be used to construct family trees, identify unknown relatives, and uncover previously unknown connections between families. In addition, IBD segment analysis can help researchers to overcome some of the challenges associated with traditional genealogy research methods, such as incomplete or inaccurate records and limited access to historical data.[7]
False positive matches and excess IBD sharing
There are some regions of the genome that are prone to excess IBD sharing, known as pile-up regions.[2] These regions can lead to false positive matches, where two individuals appear to share an IBD segment but do not actually have a recent common ancestor. Additionally, the boundaries of IBD segments can be somewhat fuzzy, leading to uncertainty in the exact locations of the segment boundaries.[2] Researchers must then take these regions into account when analyzing IBD data to avoid drawing incorrect conclusions about relationships between individuals. There are some populations that are known to have higher levels of IBD than others due to factors such as geographic isolation and small population sizes, which can also lead to false positive matches if not taken into consideration during analysis. Despite the challenges associated with pile-up regions, the use of IBD segments in genealogical research has led to many discoveries and breakthroughs in recent years; for example, by identifying IBD segments among individuals with the same rare genetic disorder, researchers have been able to pinpoint the specific genetic mutation responsible for that disorder.[1] This information can then be used to develop targeted therapies and treatments.
See also
Explore more about identity by descent
- The MyHeritage DNA test
- Shared DNA: How Much DNA Do You Share with Your Relatives?, article by Daniella Levy on the MyHeritage Knowledge Base
- Evaluating Shared DNA, webinar by Paul Woodbury on Legacy Family Tree Webinars
References
- ↑ 1.0 1.1 1.2 1.3 Sticca, Evan L.; Belbin, Gillian M.; Gignoux, Christopher R. Current Developments in Detection of Identity-by-Descent Methods and Applications. Front Genet. 2021; 12: 722602.
- ↑ 2.0 2.1 2.2 2.3 Identical by descent. International Society of Genetic Genealogy Wiki
- ↑ Thompson, Elizabeth A. Identity by Descent: Variation in Meiosis, Across Genomes, and in Populations. Genetics. June 2013; 194(2): 301–326.
- ↑ Ralph, P.; Coop, G. The Geography of Recent Genetic Ancestry across Europe. PLOS Biology. 11
- ↑ 5.0 5.1 Gusev,Alexander; Palamara, Pier Francesco; Aponte, Gregory; Zhuang, Zhong; Darvasi, Ariel; Gregersen, Peter; Pe'er, Itsik. The architecture of long-range haplotypes shared within and across populations. Molecular Biology and Evolution, 29(2), 473-486. February 2012.
- ↑ Li, Hong; Glusman, Gustavo; Hu, Hao; Shankaracharya,Caballero, Juan; Hubley, Robert; Witherspoon, David; Guthery, Stephen L.; Mauldin, Denise E.; Jorde, Lynn B.; Hood, Leroy; Roach, Jared C.; Huff, Chad D. Relationship estimation from whole-genome sequence data. PLoS Genetics, 10(1), e1004144. January 30, 2014
- ↑ Abel, Sarah; Frieman, Catherine. On gene-ealogy: identity, descent, and affiliation in the era of home DNA testing. The Anthropological Society of Nippon, 2023.


